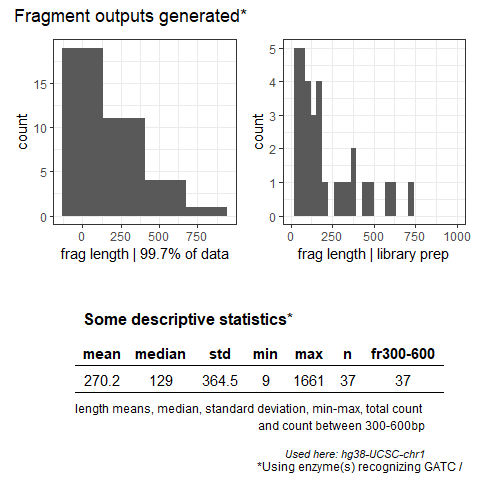
The goal of FROGmentator is to calculate fragment lengths upon template DNA fragmentation with specific pattern recognizing, (restriction) enzymes
Installation
You can install the development version of FROGmentator from GitHub with:
# install.packages("devtools")
devtools::install_github("pauliusbaltrusis/FROGmentator")