Amplitude Multiplexing
Amplitude multiplexing is a Bio-Rad’s droplet digital PCR strategy that leverages a single fluorophore to detect multiple targets by varying the concentrations of the primers and probes or the sizes of the amplicons (EvaGreen). This approach allows to distinguish between different targets based on the fluorescence intensity, which correlates with the final concentration of the primer-probe(s) or amplicon length generated and present in each droplet. By designing assays with distinct primer-probe concentrations or amplicon lengths, multiple droplet clusters can be generated that emit varying fluorescence levels, effectively creating a “multiplex” environment despite using the same dye. A variant of amplitude multiplexing - radial multiplexing - is a yet another strategy and can be utilized to multiplex targets along the diagonal, by shifting the clusters to the right (if already using a FAM probe). This shifting of the FAM cluster to the right is achieved by simply spiking in the same probe sequence but labelled with a HEX fluorophore instead. Thus, the ratio of FAM:HEX labelled probe will uniquely determine the position of the generated cluster and can further enhance the multiplexing potential.
The amplitude multiplexing method not only simplifies the experimental setup (and in turn bypasses the need for high-tier multiplexing systems with many detection channels) by reducing the number of fluorescent dyes needed but also enhances throughput and reduces costs. Amplitude multiplexing is particularly beneficial in complex analyses, such as detecting various mutations or gene copy numbers in a single reaction, enabling a more comprehensive understanding of genetic variation while maintaining the high sensitivity and precision that ddPCR is known for.
Examples below showcase amplitude multiplexing for:
-
Gene target detection & quantification (technical example):
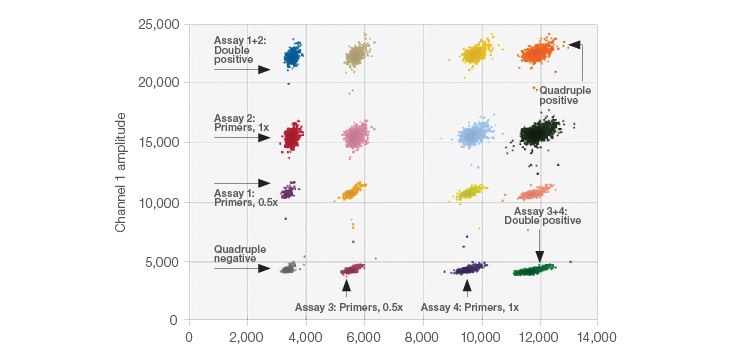 source: Bio-Rad
source: Bio-Rad -
Multiplex detection of α- and β-tryptases as well AP3B1 reference gene
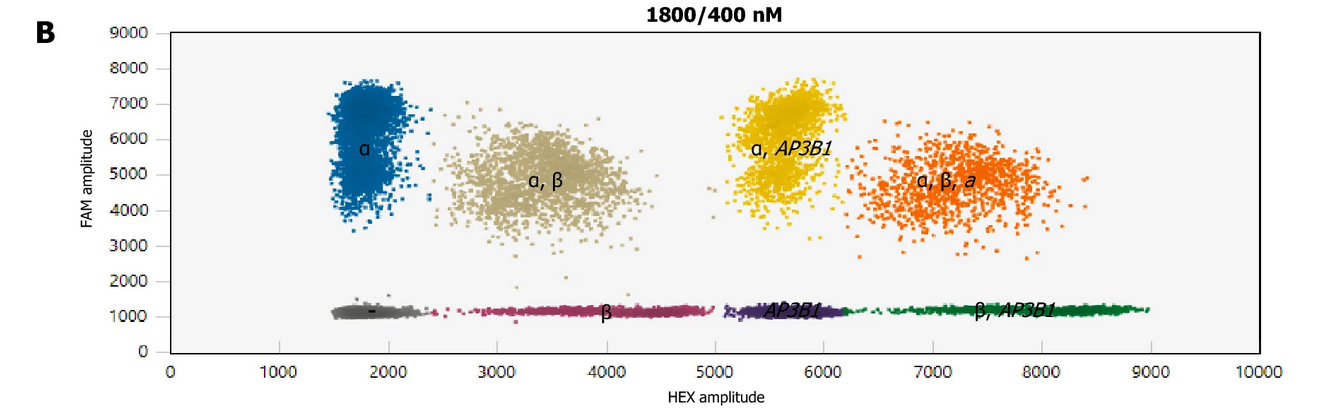
source: Svetina et al., 2024, Figure 2B -
Multiplex detection of multiple KRAS mutations in the same channel:
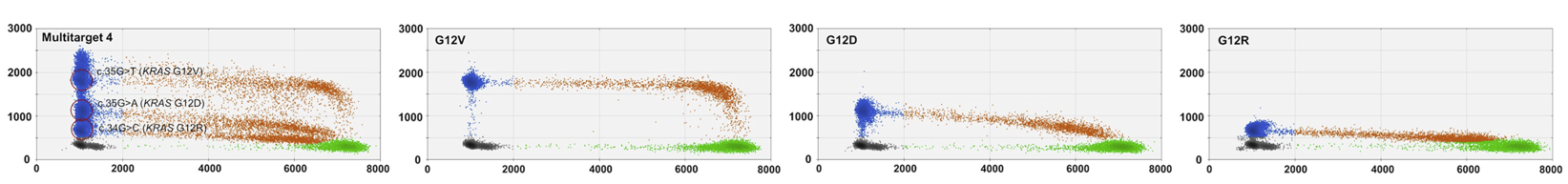 source: Hussung et al., 2020, Figure 2A
source: Hussung et al., 2020, Figure 2A -
Multiplex detection of multiple PIK3CA mutations via amplitude multiplexing and radial multiplexing (i.e. detection of a target in between the two channels):
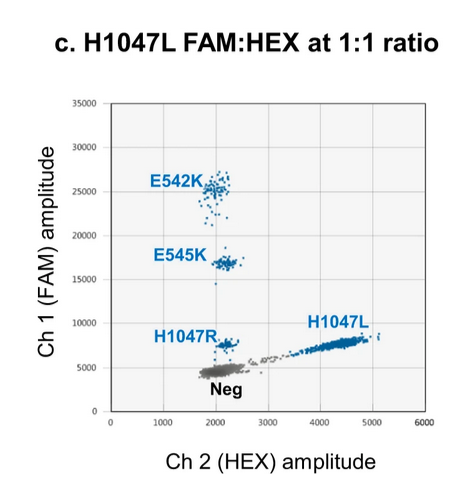
source: Rowlands et al., 2019, Figure 7C
Thus by adjusting primer-probe concentrations, amplicon lengths (if working with EvaGreen) or spiking in a fraction of your probe labelled with a different fluorophore (e.g., HEX) one can multiplex and detect multiple target sequences either within the same channel or in between the two channels.
To find out more about how to optimize such assays, check out Optimization.