Locked-Nucleic Acids & Degenerate Bases
Locked Nucleic Acids (LNAs) and degenerative bases are important tools in designing primers and probes for digital droplet PCR (ddPCR), especially when targeting difficult regions of the genome. LNAs are chemically modified nucleotides where the ribose ring is locked in a rigid conformation, which significantly increases the thermal stability of the DNA duplex. Incorporating LNAs into primers or probes enhances their binding affinity to complementary sequences, allowing for more precise and stable hybridization, even under challenging conditions such as high GC content, secondary structures, or regions with low sequence complexity. This increased stability can result in improved specificity and sensitivity in ddPCR assays, reducing the risk of non-specific binding or amplification, which is particularly valuable when detecting rare mutations or small genetic variations (i.e. SNPs, especially transitions). Further reading material on LNAs can be found here.
In addition, the use of degenerative bases (or degenerate bases), which represent mixtures of possible nucleotides at specific positions within primers or probes, can be helpful when targeting sequences with polymorphisms or unknown variations (see the image below). Degenerative bases allow primers and probes to bind to multiple sequence variants without the need for designing multiple different oligonucleotides. This flexibility can be especially useful when working in regions of the genome with high variability or when exact sequence information is incomplete or uncertain. By incorporating degenerative bases, ddPCR assays can effectively capture a broader range of target variants, ensuring that key mutations or variants are not missed due to sequence mismatches. You can find more information on degenerative bases on IDTs website.
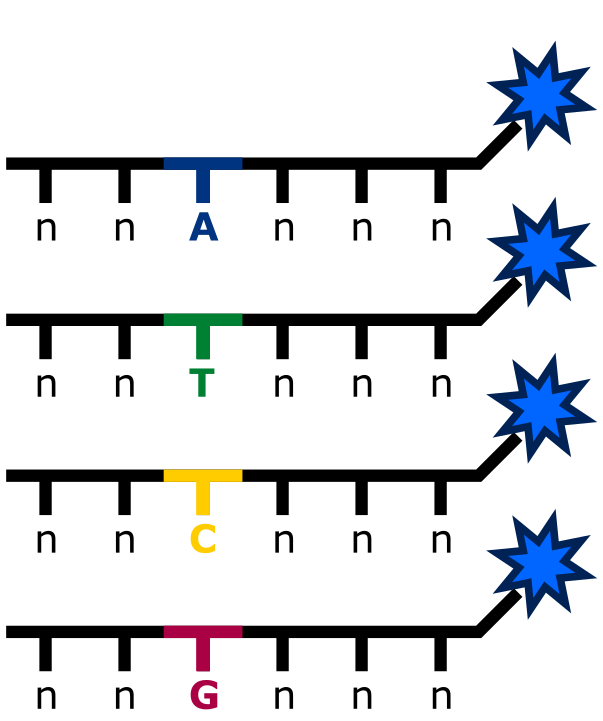
An example of a probe (shown without a quencher) with a degenerate base, intended to target loci exhibiting variability (eg., SNPs)
Together, LNAs and degenerative bases enhance the robustness of ddPCR assays by improving primer and probe binding efficiency, particularly in difficult-to-target regions. This enables more reliable and sensitive detection of genetic variants, even in complex genomic environments where traditional primers or probes might struggle.