Drop-off Probes
When working with SNPs or precise genome editing methods like homology-directed repair (HDR), using multiple probes can be advantageous. One probe is designed to target the wild-type sequence, while another targets the mutant sequence (see part A in the image below).
For applications involving non-homologous end joining (NHEJ) events—frequently resulting from CRISPR/Cas9-induced double-strand breaks (DSBs)—drop-off probes are particularly useful in droplet digital PCR (ddPCR). These probes are designed to detect small insertions or deletions (indels) created at the break site during the repair process.
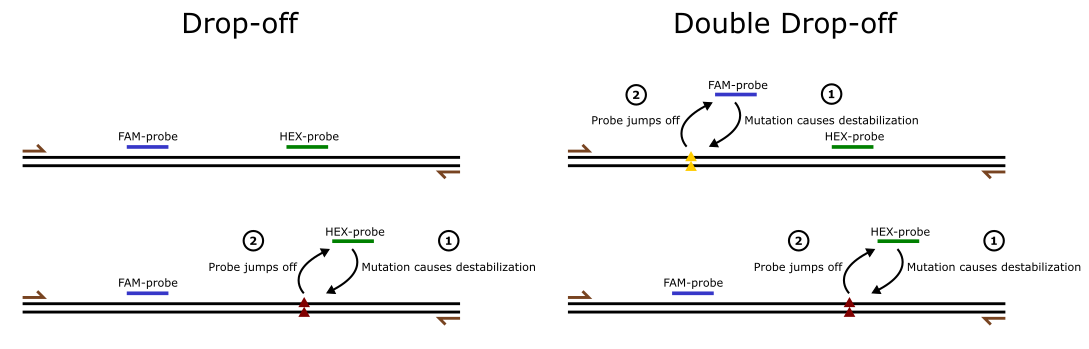
A drop-off probe typically binds to the wild-type sequence adjacent to the CRISPR cut site (see part B in the image below). This probe is labeled with a fluorophore and emits a signal when hybridized perfectly to the intact wild-type sequence. However, if the sequence is disrupted by an indel, the probe can no longer bind effectively, resulting in a “drop-off” in fluorescence.
In ddPCR, DNA molecules are partitioned into thousands of droplets, each containing zero, one, or a few DNA molecules. Some droplets will contain unmodified wild-type sequences, while others will contain sequences with indels. By measuring fluorescence in each droplet, ddPCR enables highly sensitive detection of CRISPR-induced mutations, quantifying the proportion of modified DNA in a sample.
Double drop-off probes in ddPCR are an extension of the single drop-off probe method, allowing for the detection of two different mutations or events, only one of which occurs at any given time in a particular genomic region (see part C in the image below). This method is particularly useful when working with cases where two possible outcomes or mutations may arise but are mutually exclusive.
Double drop-off probes extend the single drop-off method, enabling the detection of two different mutations or events that are mutually exclusive (see part C in the image below). This approach is particularly useful in scenarios where two possible outcomes arise in the same genomic region, but only one occurs in any given event.
In a double drop-off assay:
- Two distinct probes are designed to target separate sequences or regions, each representing a potential mutation or genetic event.
- The probes are labeled with different fluorophores, allowing for simultaneous detection.
- When a mutation disrupts one target site, its corresponding probe cannot bind, leading to a “drop-off” in signal for that probe. If the second mutation is absent, the second probe will continue to emit its signal, indicating the mutation’s exclusivity.
This setup is particularly useful in applications like:
- Detecting allelic dropout, where one allele undergoes a mutation while the other remains intact.
- Gene editing experiments where two distinct outcomes are possible, and the assay discriminates between them in a single reaction.
In the example below, you can see a visualization of a typical a single drop-off assay with a drop-off probe and double drop-off assay for simultaneous detection of two mutually exclusive mutations.
 Single Drop-off assay vs Double Drop-off assay
Single Drop-off assay vs Double Drop-off assay