Mutation-Selected Amplification ddPCR
Mutation-Selected Amplification droplet digital PCR (MSA-ddPCR), a technique that reminds one of allele-specific digital PCR, can be used to determine the SNP status of a target locus while simultaneously amplifying a universal reference locus. Hu et al., 2024 developed this method and used it to study the TP53R249S substitution. Their assay demonstrated greater sensitivity than amplicon-based high-throughput sequencing, reliably detecting mutation frequencies as low as 0.01%.
However, the practical differences between MSA-ddPCR and more traditional ddPCR SNP detection approaches—such as allele-specific ddPCR (where the 3’ end of the primer is blocked by the presence of a SNP) or assays using two probes with single nucleotide differences—are unclear. For instance, regular mutation detection assays using competing probes, like those described by Dong et al., 2018, achieve comparable limits of detection (LOD). Moreover, MSA-ddPCR introduces additional complexity, requiring multiple primer pairs and essentially two rounds of template amplification. A reasonable concern could thus be that the initial amplification step uses an oligo with a long unbound tail, which could affect downstream reaction efficiency (and therefore the droplet cluster separation).
Another important limitation is that MSA-ddPCR does not measure a “true” allele frequency. The use of different primer pairs for the variant-specific and universal loci could introduce subtle variability due to differences in primer efficiency between the two amplicons.
Despite these limitations, MSA-ddPCR represents a novel approach and is effective for detecting low-frequency variants, as demonstrated by its sensitivity and low LOD.
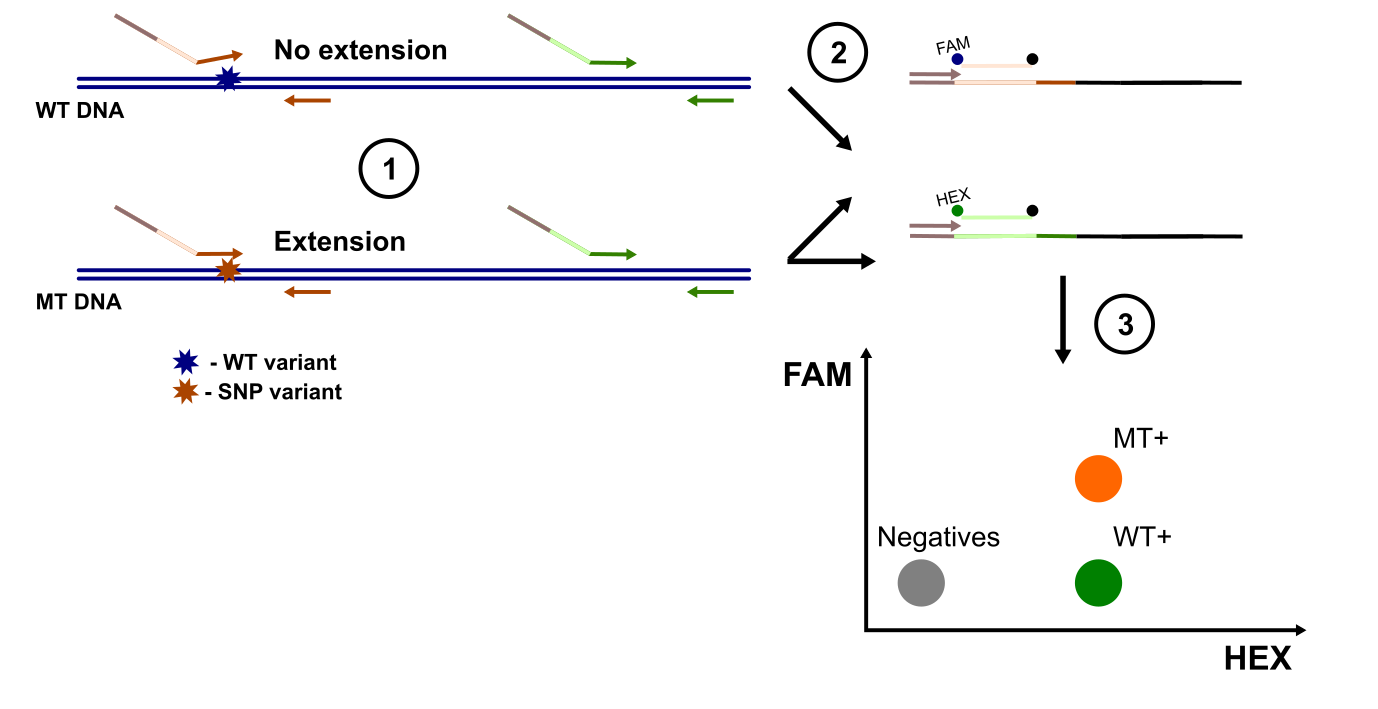
The principle behind MSA-ddPCR as reported by Hu et al., 2024