Linkage and Phasing
By focusing on the presence or absence of abnormal numbers of double-positive droplets, ddPCR can be used to investigate linkage and even phase multiple variants at different loci. A study by Regan et al., 2015 outlines one such approach, which evaluates molecular integrity and uses restriction enzymes to test linkage.
In the example below:
(a) When variants are unlinked and located on separate homologous chromosomes, random partitioning during droplet generation produces fewer double-positive droplets for each allele.
(b) If variants A and T are linked (i.e., on the same homologous chromosome and in close proximity), the number of double-positive droplets will be abnormally higher than expected.
(c) To confirm whether A and T are truly linked, the sample can be treated with a restriction enzyme that cuts the DNA between A and T. This digestion disrupts the physical linkage, resulting in significantly fewer double-positive droplets, thus validating the suspected linkage.
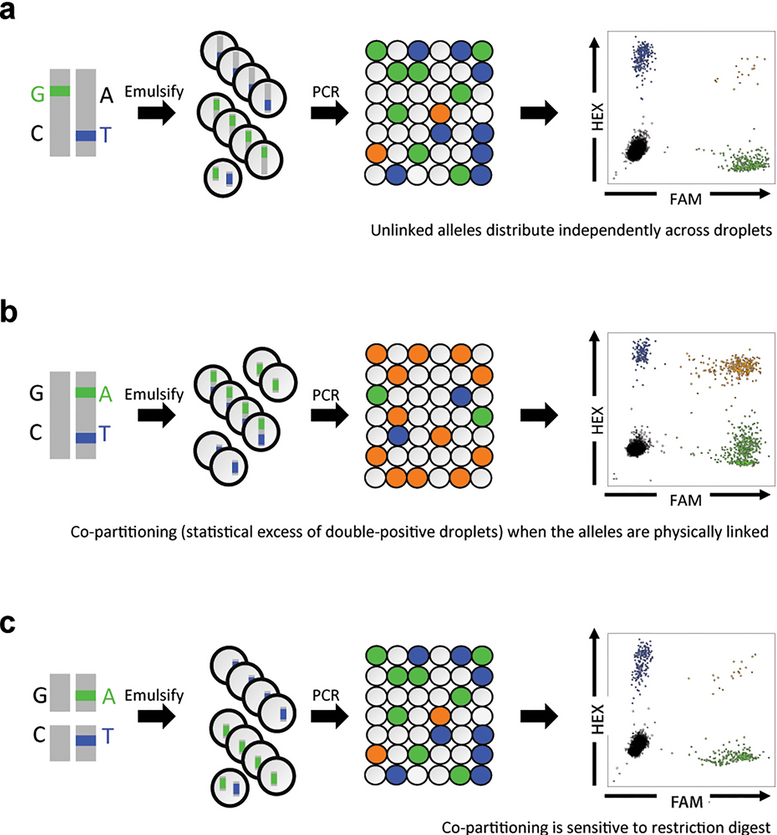 source: Regan et al., 2015
source: Regan et al., 2015
Thus, using restriction enzymes, we can test for the existence of linkage between genetic variants by disrupting their physical connection. If the variants are linked, treating the sample with a restriction enzyme that cuts between them will reduce the number of double-positive droplets in ddPCR, confirming their prior linkage.