Mutation Detection with Amplitude Multiplexing
If your goal is to identify and quantify several mutations using a system like the QX200 (2-channels), a more specific approach is necessary. In amplitude multiplexing, the detection of multiple targets relies on differences in fluorescence intensity (amplitude) within the same channel. So, probes for different mutations can be labeled with the same fluorophore, with each probe’s interaction with its target mutation generating droplets of distinct fluorescence amplitudes. These differences allow the discrimination of mutations in 1D or 2D plots.
Example 1: KRAS Mutation Detection
Consider the detection of three KRAS mutations (G12V, G12D, G12R) in one well using optimized primer/probe concentrations:
- G12V assay: 900 nM primers, 250 nM probe
- G12D & G12R assays: 450 nM primers, 125 nM probe each
As shown in a study by Hussung et al., 2020, this setup enables the simultaneous detection of all three mutations by clustering droplets based on fluorescence intensity.
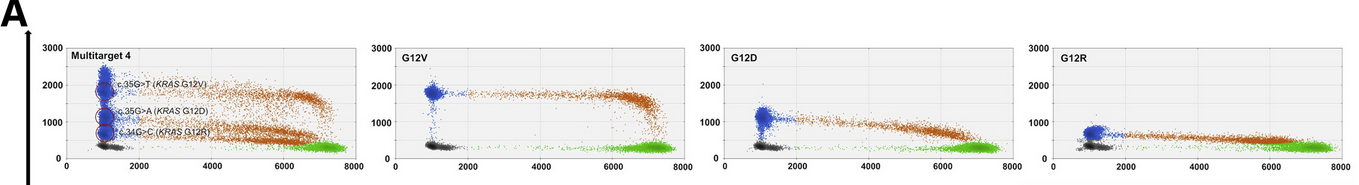 source: Hussung et al., 2020, Figure 2A
source: Hussung et al., 2020, Figure 2A
Example 2: Radial Clustering with Mixed Channels
Rowlands et al., 2019 demonstrated a novel approach using both FAM and HEX channels for mutation detection. For instance, the PIK3CA H1047L mutation was detected using probes labeled with FAM or HEX at varying ratios (e.g., 3:1 or 1:1), resulting in droplet clusters positioned along the diagonal between the two channels . This technique expands the multiplexing capability by exploiting inter-channel fluorescence.
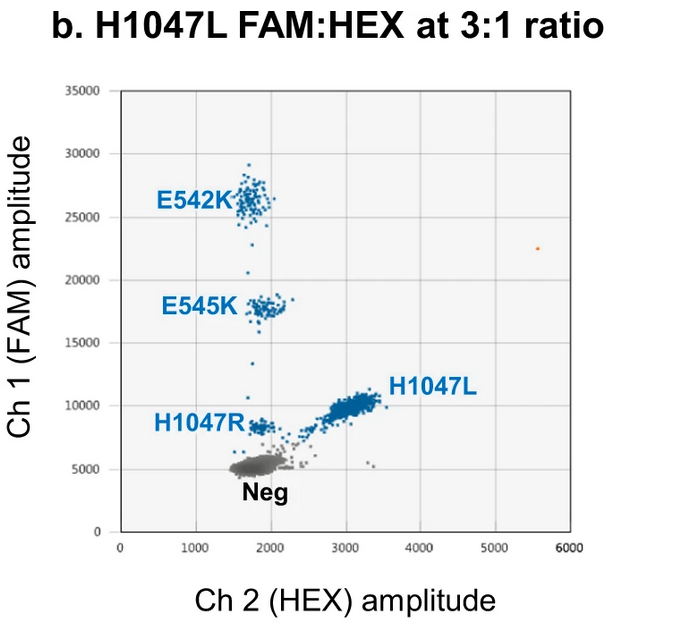 source:Rowlands et al., 2019, Figure 7b
source:Rowlands et al., 2019, Figure 7b
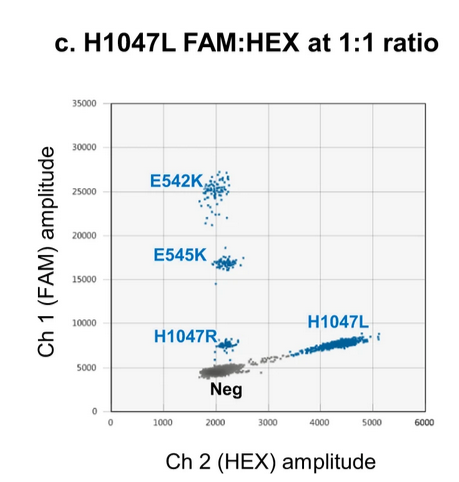
source:Rowlands et al., 2019, Figure 7c
Example 3: Multiplex Drop-Off Probe Assays
If specific mutation types aren’t critical, a multiplex drop-off probe assay might be suitable for identifying mutations across multiple hotspot loci. Yu et al., 2023 showcased a method combining drop-off assays with SNP probes to detect mutations in KRAS, NRAS, and BRAF exons known for mutation hotspots (Figure 1). This approach provides broad mutation coverage in a streamlined setup.
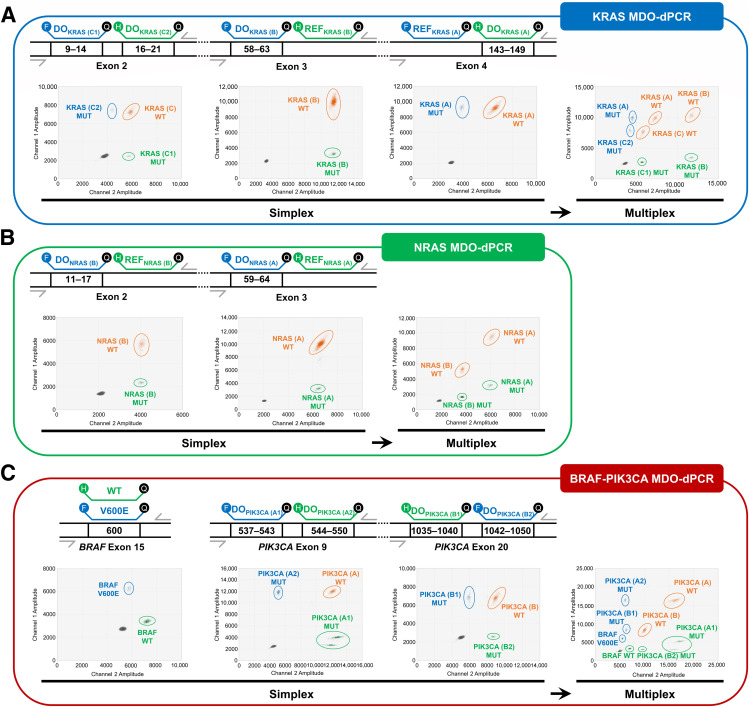 source: Yu et al., 2023, Figure 1
source: Yu et al., 2023, Figure 1
Some Considerations
While multiplexing increases efficiency, it also raises complexity. Overlapping fluorescence signals can result in messy 2D plots if reactions are poorly optimized. To avoid this:
- Optimize individual assays before multiplexing.
- Benchmark each reaction to ensure clear droplet clustering.
- Gradually combine assays, adjusting concentrations to maintain signal separation.
By following these guidelines, you can achieve robust and accurate multiplex mutation detection.