T-Blocker qPCR
The paper by Kim et al., 2018 introduces the T-blocker, a probe-free quantitative PCR (qPCR) assay designed to detect somatic mutations with a sensitivity down to 0.1% mutant allele frequency. This method enhances the traditional allele-specific PCR by incorporating a T-blocker—a specially designed oligonucleotide that suppresses wild-type allele amplification, thereby increasing the specificity for mutant alleles - in a similar fashion to BDA. Unlike conventional techniques that rely on dye-labeled probes, the T-blocker approach utilizes intercalating dye-based qPCR chemistry (eg., SYBR Green), simplifying the assay design and reducing costs.
Kim et al., developed four assays targeting specific KRAS and BRAF mutations to demonstrate the efficacy of the T-blocker method. By employing a customized fast PCR protocol, they achieved consistent and highly sensitive detection of mutant alleles amidst a predominant wild-type background. The results indicate that the T-blocker assay provides a simple, robust, and cost-effective alternative for detecting low-frequency somatic mutations, which is crucial for accurate molecular diagnostics in personalized cancer treatment.
The image below summarizes the outcomes of running both WT and MT template mixtures with a T-Blocker: In A) and C) the WT template amplification is either blocked or severely dampened due to the mismatch between the primer and the template. in B) The primer for the MT template binds to the MT (i.e., SNP containing) template and thus the polymerase displaces the T-Blocker during polymerization.
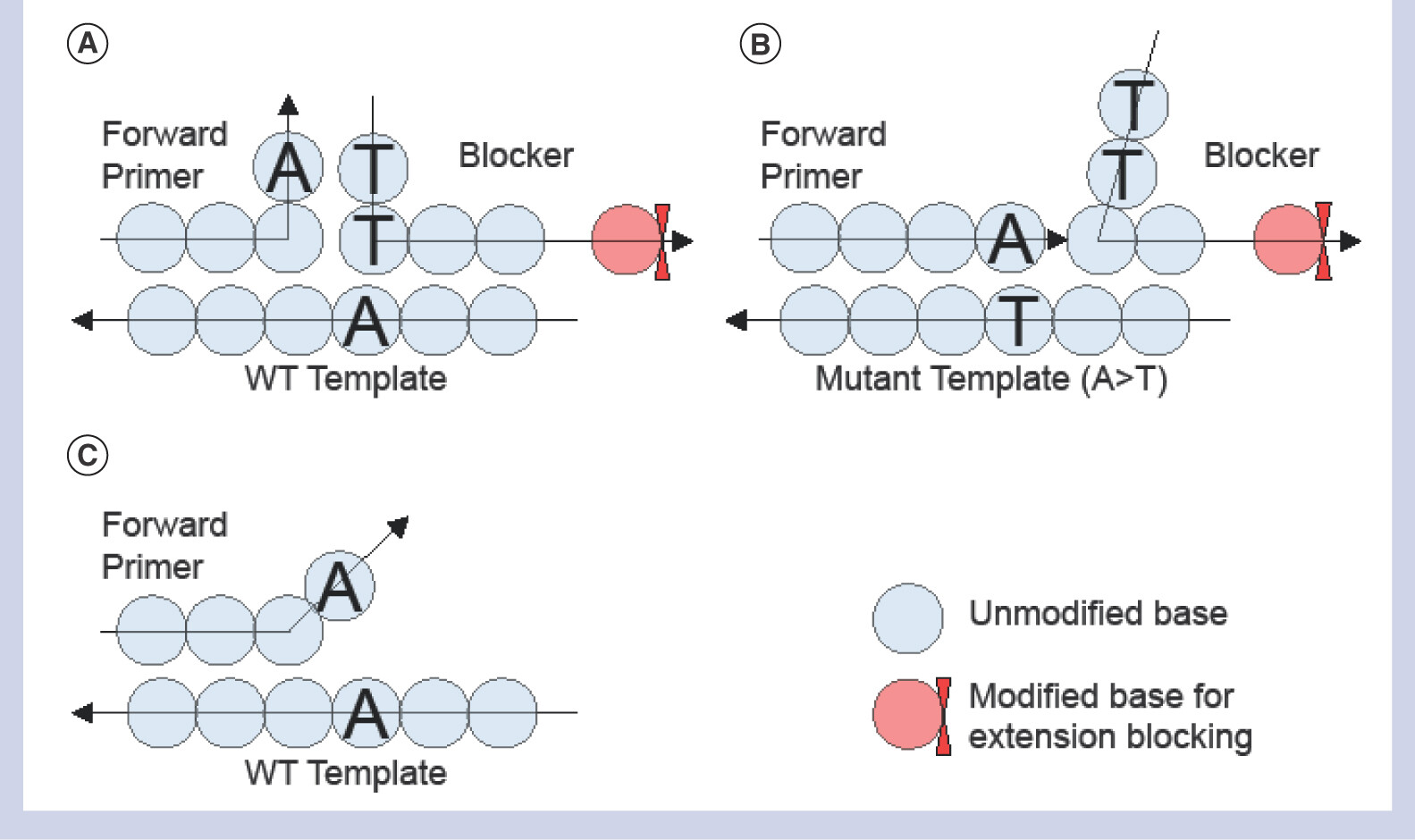 source: Kim et al., 2018, Figure 1
source: Kim et al., 2018, Figure 1