Dark Probes
Dark probes are highly effective in eliminating the signal from wild-type sequences by binding specifically to the wild-type target and quenching any potential fluorescence (see the image below). Since they lack a fluorescent reporter, these probes do not emit a signal upon hybridization. This effectively “silences” the wild-type allele, allowing the fluorescent signal from the mutant-specific probe to stand out more clearly. As a result, the overall background noise from the wild-type sequence is minimized, leading to highly specific detection of the mutant allele, even when present at very low frequencies. This specificity is critical in applications where detecting rare mutations in a large background of wild-type DNA is necessary, such as in liquid biopsies for cancer or detecting drug-resistant mutations.
Keep in mind that the trade-off of using a dark probe to “silence” the wild-type signal is that the wild-type allele is not detected (and thus not quantified), ultimately preventing one from calculating the frequency of the mutant allele in sample(s). Thus, this approach is not suitable for situations where both wild-type and mutant signals need to be quantified. Since dark probes eliminate the wild-type signal, they obscure information about the copy number or presence of the wild-type allele, making it impossible to measure both alleles in the same reaction. For applications requiring quantification of both wild-type and mutant sequences - a different strategy, e.g. using dual-labeled probes with distinct fluorophores for each allele, would be necessary. These alternative probes would allow the simultaneous detection and quantification of both wild-type and mutant signals.
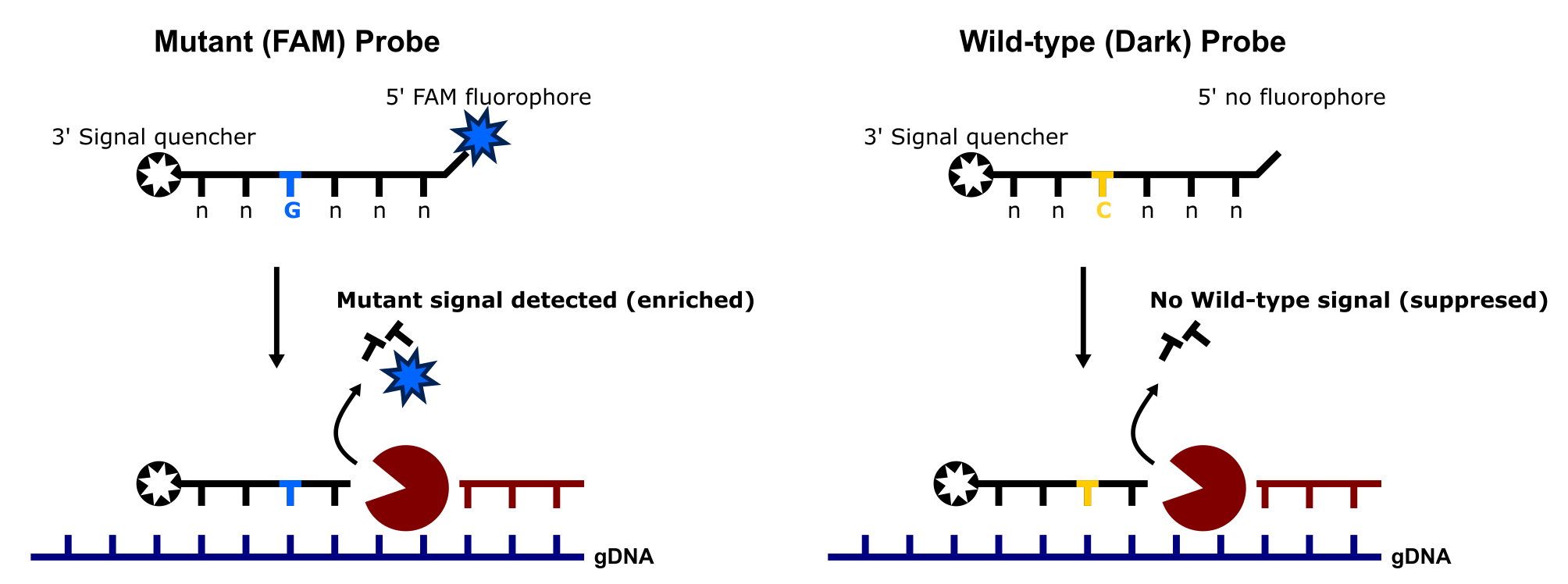 An example of how the fluorescence signal from the wild-type template is suppressed through the use of a dark probe.
An example of how the fluorescence signal from the wild-type template is suppressed through the use of a dark probe.