Simplified Multiplexing Strategies for Mutation Detection
When detecting multiple mutations, the conventional approach involves designing mutation-specific probes, each tagged with a distinct fluorophore (e.g., FAM, HEX). These probes are engineered to bind uniquely to sequences with specific mutations or wild-type alleles. In a single ddPCR reaction, different fluorophore signals correspond to different mutations. However, this method is limited by the number of fluorescent channels available in the ddPCR system.
If the goal is to identify whether any mutation is present without specifying which one, a simpler approach can be used:
Multiple Probes with the Same Fluorophore: Design several probes (closely located within the same amplicon and flanked by a single primer pair) targeting different mutations. All probes are labeled with the same fluorophore (e.g., FAM). In this setup, a signal in the fluorophore’s channel indicates the presence of at least one mutation, but the specific mutation responsible is not determined.
This strategy is used in Bio-Rad’s ddPCR KRAS G12/G13 Screening Kit, which detects seven SNPs in the KRAS gene at codons 12 and 13. The kit employs a set of primers and multiple probes, all labeled with FAM, to identify whether any of the SNPs are present. However, it does not distinguish between individual SNPs. The part A of the illustration below demonstrates this approach.
Another alternative is the drop-off probe strategy: The drop-off probe targets a mutation-prone hotspot and is complementary to the wild-type sequence. If a mutation disrupts the probe-binding site, the probe cannot hybridize, leading to a “drop-off” in fluorescence. A positive signal indicates the absence of mutations, while a drop-off suggests the presence of one or more mutations in the hotspot.
This approach requires only one fluorophore and is particularly effective for screening mutation-prone regions. An example is shown in B.
Both strategies enable efficient screening for mutations in a hotspot without the need for:
- Extensive optimization to differentiate fluorescence clusters for each mutation.
-
Running multiple reactions with distinct primer-probe sets.
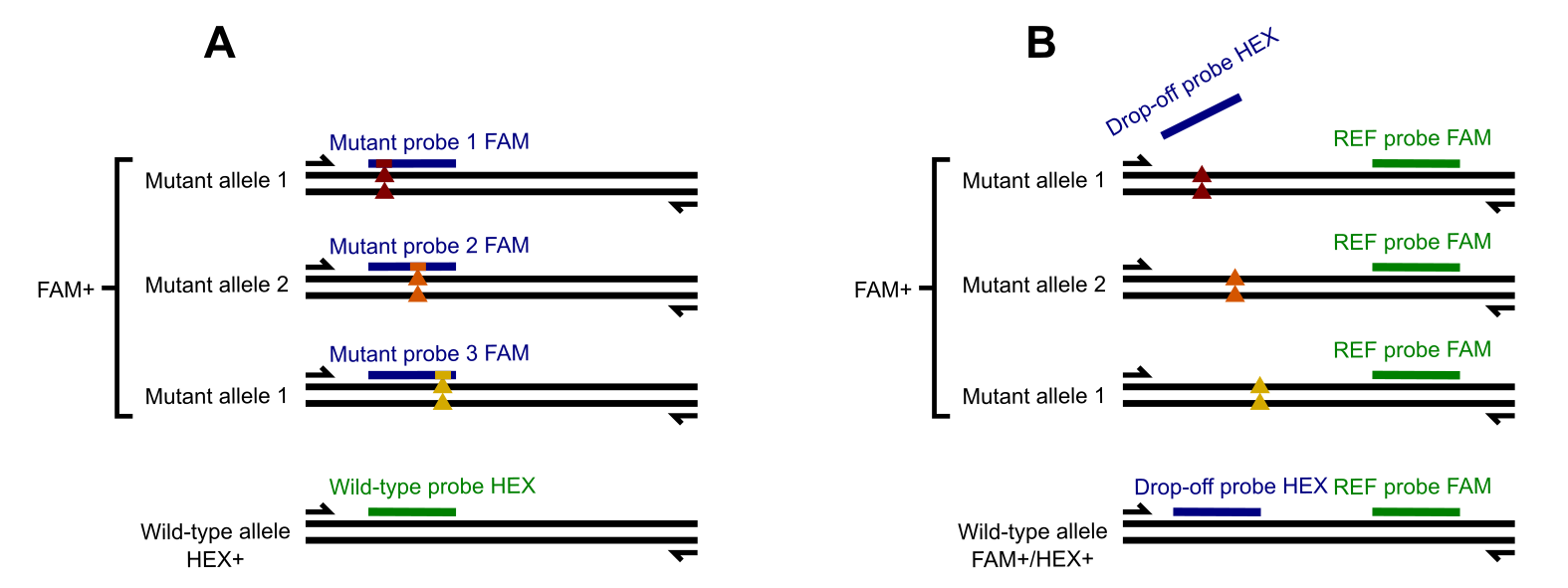 Multiple SNP detection with FAM(only)-labelled probes vs a universal Drop-off probe
Multiple SNP detection with FAM(only)-labelled probes vs a universal Drop-off probe
These methods are ideal for identifying the presence of any mutation in a hotspot when the specific identity of the mutation is less critical, such as in initial screening workflows. For a reference study, check out Decraene et al., 2018