Gene Linkage: EHEC example
A study investigating gene linkage in prokaryotes, specifically E. coli, was conducted by McMahon et al., 2017. The research focused on quantifying the linkage between the bacterial genes eae and stx, both of which must be present in the same cell to classify the strain as pathogenic EHEC. The challenge arises because commensal, non-pathogenic E. coli may harbor either of these genes independently, leading to both genes being detected in mixed populations. The key objective was to differentiate samples positive for EHEC (containing both genes within the same genome) from non-pathogenic mixed flora, where the genes are not linked.
The authors addressed this by encapsulating live E. coli cells in droplets, avoiding the analysis of gDNA mixtures. Their concept was that samples containing EHEC would produce a higher number of double-positive droplets compared to samples with only commensal E. coli, where the genes are unlinked (see image below).
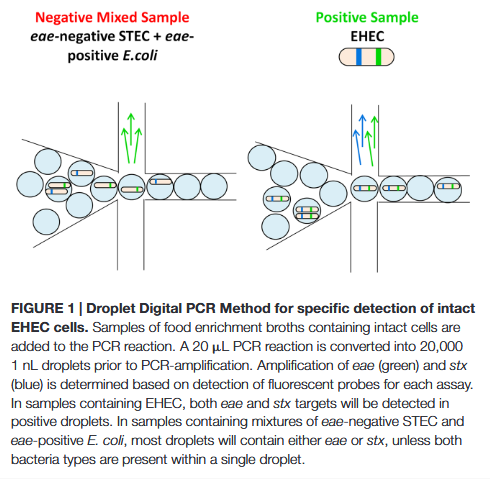
source: McMahon et al., 2017
The study revealed that samples containing EHEC consistently showed higher linkage rates between the two target genes compared to commensal E. coli samples, even at extreme dilutions. Remarkably, the method could distinguish a sample containing just one EHEC cell among 10,000 background E. coli cells, despite significant background microbiota.