Single-Cell Transcript Profiling & Radial Multiplexing
A poster by Claudia Litterst et al., 2014 outlines a highly efficient workflow for single-cell transcript quantification using droplet digital PCR (ddPCR). This method is particularly advantageous for mRNA biologists aiming to quantify transcripts without the need for pre-amplification of cDNA.
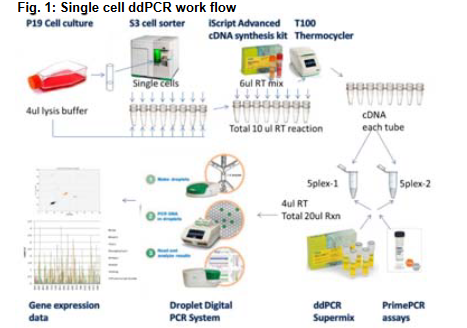
source: Claudia Litterst et al., 2014
Workflow Summary
- Cell Sorting and Lysis:
- Single cells were isolated using the S3 Cell Sorter and resuspended in 4 µL of lysis buffer containing IDTE (pH 8), RNase inhibitor, and 0.1% Triton-X100.
- cDNA Synthesis:
- An additional 6 µL of the iScript Advanced RT cDNA synthesis kit supermix was added to the lysate.
- The total 10 µL reaction was incubated in a thermocycler for reverse transcription.
- Partitioning for ddPCR Reactions:
- The resulting cDNA was divided into two 4 µL aliquots, each used for ddPCR in separate reactions (referred to as 5plex 1 and 5plex 2).
- Each aliquot targeted five distinct mRNA transcripts, achieving detection of 10 unique targets per sample across two wells.
Most importantly, the method bypasses cDNA pre-amplification, reducing bias and preserving the original transcript ratios.
Key Feature: Radial Multiplexing
The study introduces a clever amplitude-based multiplexing strategy called radial multiplexing. This method involves:
- ddPCR Assays:
- Two identical assays were designed for each target, differing only in the fluorescent labels on the probes:
- One assay used a FAM-labeled probe.
- The other used a HEX-labeled probe.
- Both assays shared identical primer and probe sequences.
- Two identical assays were designed for each target, differing only in the fluorescent labels on the probes:
- Probe Ratios:
- FAM and HEX assays were combined at different ratios for each target, creating unique signal clusters for individual transcripts.
- 2D Clustering:
- By leveraging the 2D space (X = channel 1, Y = channel 2), the method allows each target to form a distinct cluster on the plot.
Probe Ratio Table The table shows the mixing ratios of FAM and HEX probes used in order to generate distinct clusters on the radial. 5 targets were detected in each 5plex reaction by reducing the concentration of the FAM (%) probe (100%->75->50->25->0) while simultaneously increasing the concentration of the HEX (%) probe (0->25->50-75->100%)
| Gene | FAM (%) | HEX (%) | Assay Volume (µL) |
|---|---|---|---|
| 1, 6 | 100 | 0 | 1.5 |
| 2, 7 | 75 | 25 | 1.0 |
| 3, 8 | 50 | 50 | 1.0 |
| 4, 9 | 25 | 75 | 1.0 |
| 5, 10 | 0 | 100 | 1.5 |
Results and Interpretation
The culmination of this approach is the generation of a 2D plot, where each of the 10 targets is detected and visualized as a distinct cluster. By running the same sample across two wells on the QX200 ddPCR system, researchers can confidently quantify all 10 targets with high precision.
This workflow represents a significant advancement in single-cell transcript profiling, offering a robust and ultra-sensitive method for multiplexed transcript detection without introducing amplification biases.
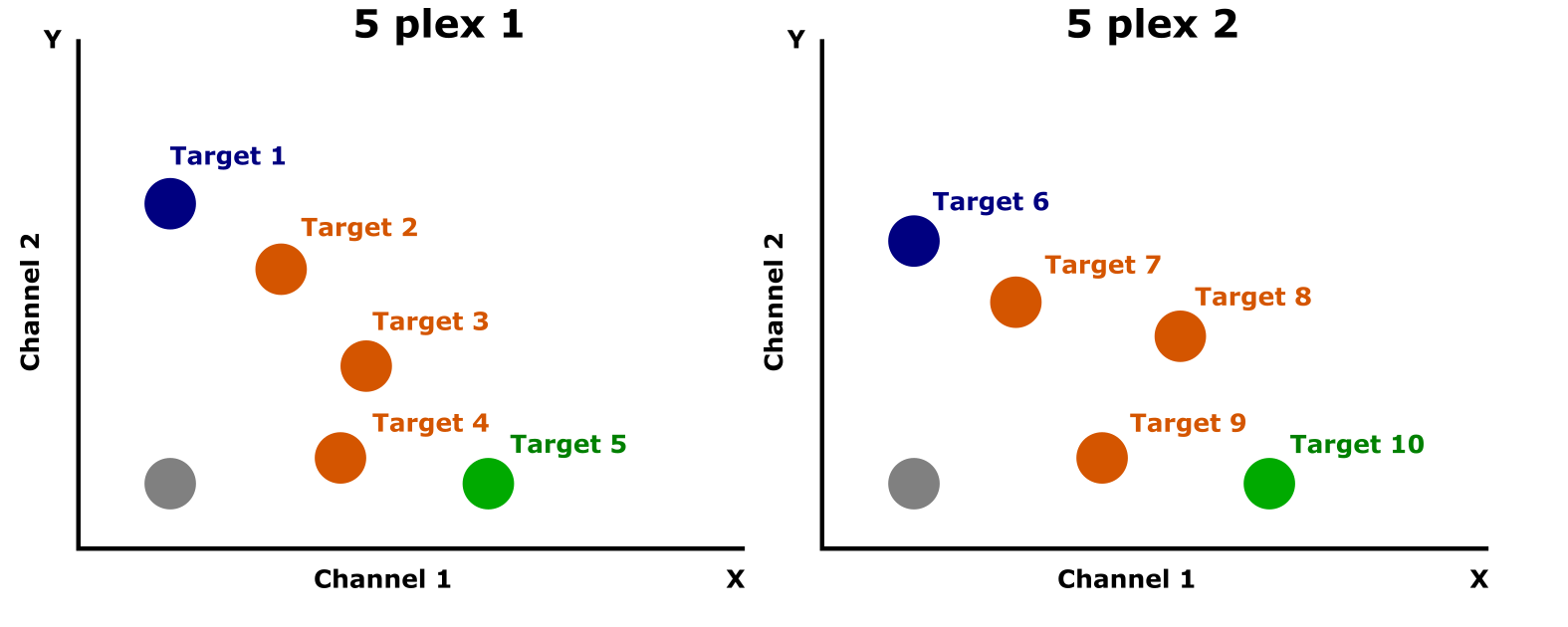 A simplified visual representation of the different target-containing droplet clusters detected (5 +5) in the 5plex 1 & 2 duplicates and their unique distributions between the 2 (FAM & HEX) channels.
A simplified visual representation of the different target-containing droplet clusters detected (5 +5) in the 5plex 1 & 2 duplicates and their unique distributions between the 2 (FAM & HEX) channels.