RPA
RPA is a DNA amplification technique designed for rapid, sensitive, and specific nucleic acid detection under isothermal conditions (similar to LAMP), avoiding the need for thermal cycling. Initially invented by Piepenburg et al., 2006, its distinguishing features are:
Principle of Amplification:
- RPA uses a recombinase-primer complex to facilitate homologous pairing of primers with complementary target DNA sequences.
- The recombinase binds primers and scans the target DNA for homologous sequences, enabling strand exchange and primer binding.
Mechanism:
- Recombinase Proteins: Form a filament with primers to guide them to their complementary sequences on the target DNA.
- Single-Strand Binding (SSB) Proteins: Stabilize the displaced single strand during primer binding.
- DNA Polymerase: Extends the primers and displaces the downstream strand, enabling continuous amplification at a constant temperature (37–42°C).
Products:
- RPA generates amplified DNA fragments in a sequence-specific manner. Amplification is exponential and can be detected in real-time or endpoint assays using fluorescent probes, lateral flow strips, or gel electrophoresis (refer to the original paper).
Advantages:
- Speed: Amplification occurs rapidly, typically within 20–40 minutes.
- Low Temperature: Operates at a constant temperature close to physiological conditions (37–42°C).
- High Sensitivity: Detects low amounts of DNA, often in the range of femtograms.
- Versatility: Works well with crude samples and minimal preprocessing.
- Field Deployable: Requires minimal equipment, making it suitable for point-of-care or field-based applications.
Applications:
Initially developed for DNA detection, RPA has since been applied to various fields, including infectious disease diagnostics, environmental monitoring, and food safety testing.
DETECTR (CRISPR/Cas12a + RPA) method
DETECTR or DNA Endonuclease Targeted CRISPR Trans Reporter has been developed by Chen et al., 2019 and is an example of a spectacular method combining RPA with a 2-step CRISPR-Cas12a system to generate a sensitive, fluorescence based detection of specific dsDNA sequences.
DETECTR is performed by:
1) pre-amplifying the sequence of interest with RPA to yield more copies of the target of interest
2) Introducing Cas12a-sgRNA and a single-stranded fluorescence reporter into the reaction
3) a) Cas12a-sgRNA complex recognizes the specific DNA target sequence in cys, cuts it and b) performs indiscriminate trans cutting of single-stranded DNA (the fluorescence reporter) in the mix
Thus by leveraging RPA-mediated pre-amplification of a target DNA sequence and Cas12 promoted site-specific dsDNA cutting and non-specific ssDNA trans-cleavage, a rapid & sensitive method can be employed to detect target DNA concentrations as low as, e.g., 0.1 copies/uL in under 60 mins (see Cao et al., 2025).
The steps in the DETECTR workflow are summarized in the illustration below:
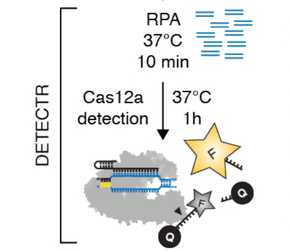
source: Chen et al., 2019, adapted from Figure 4 C