Structural Variants
Although most applications of droplet digital PCR (ddPCR) focus on detecting small genetic variations such as indels or single nucleotide polymorphisms (SNPs), the technology can also be adapted to detect and quantify large structural variations, including large excisions (up to hundreds of thousands of nucleotides) and inversions.
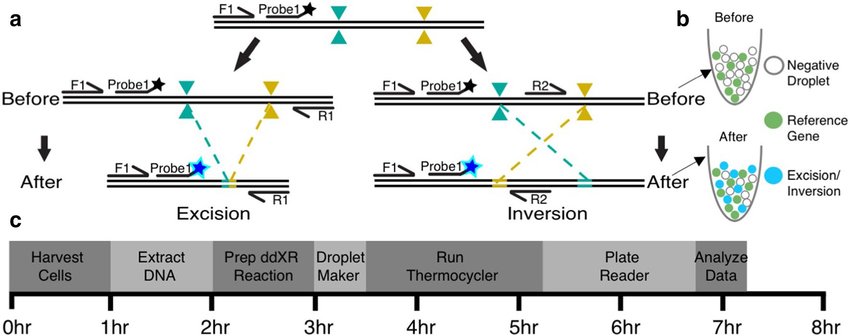
The ddXR (droplet digital PCR eXcision Reporter) method, introduced by Watry et al., 2020, is a specialized ddPCR approach designed for the precise detection and quantification of large DNA excisions and inversions, particularly those induced by gene-editing technologies such as CRISPR-Cas9. This method works by targeting sequences flanking the edited region to determine the presence or absence of the intervening sequence, thereby identifying excision or inversion events. The schematic below provides an overview of the ddXR assay.
 source: Watry et al., 2020
source: Watry et al., 2020
This is how ddXR assays work:
Probe Design: The ddXR method uses two primer pairs and a shared probe. Both pairs share a forward primer (F1) but differ in their reverse primers: one (R1) flanks the region expected to be excised, while the other (R2) spans the sequence targeted for inversion. This setup leaves the second fluorescence channel free to detect a reference gene, which serves as a baseline for normalizing structural variation frequencies.
Signal generation in the droplets: During ddPCR, DNA is partitioned into thousands of droplets, each containing zero, one, or a few DNA copies. If the target DNA remains unchanged (no excision or inversion), the primers cannot generate a complete amplicon: R1 is too distal, and R2 is in the opposite orientation. These droplets will lack fluorescence signals. However, if excision or inversion has occurred, one of the primer sets (F1R1 for excision or F1R2 for inversion) becomes functional, generating a full amplicon and a fluorescence signal in those droplets.
Quantification: By comparing the number of DNA copies exhibiting excision or inversion events to the number of reference gene copies, the method estimates the frequency of these structural variations in the sample.
The ddXR method is particularly valuable for detecting large-scale genomic edits, such as long deletions or inversions, which are often challenging to identify with traditional PCR methods. This capability is critical in therapeutic gene editing, where precise quantification of large excisions is essential for assessing editing efficiency and ensuring the accuracy of the edits.