Single-cell RNA & Protein Quantification
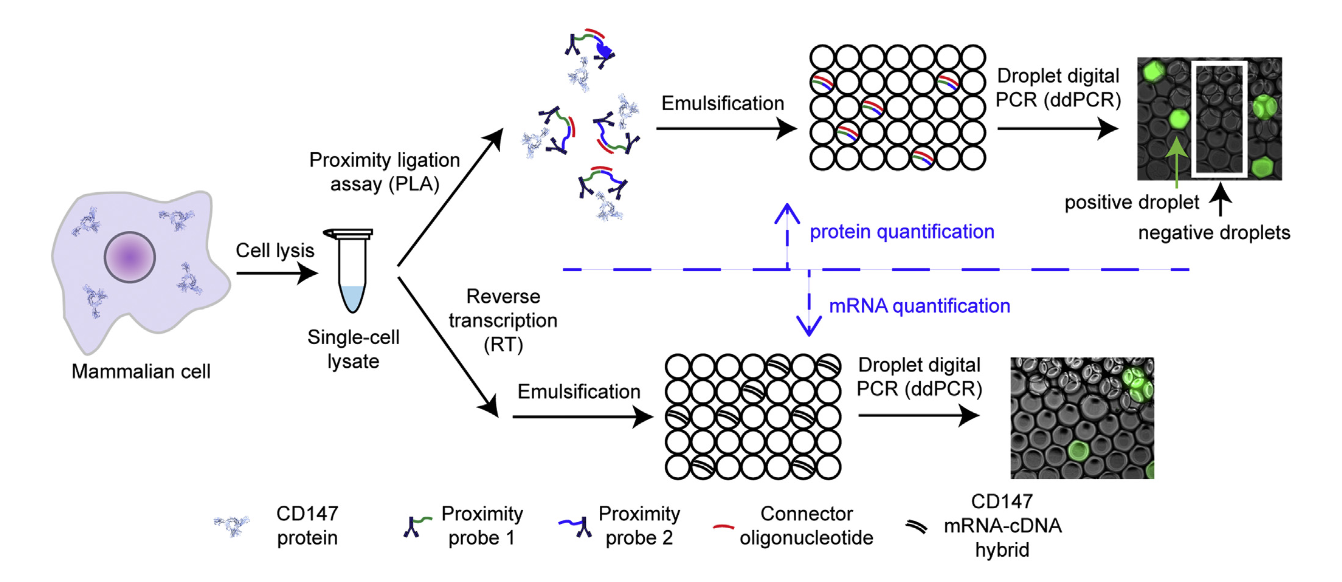
A impressive method combining digital Proximity Ligation Assay (dPLA) and RT-ddPCR was introduced by Albayrak et al. 2016. This approach enables the simultaneous and quantitative measurement of both proteins and mRNA in individual cells, offering an unparalleled level of sensitivity and resolution. By merging these two techniques, the method provides a means to track key biomolecules, which are often co-regulators of cellular function, in a highly sensitive and absolute manner.
 source: Albayrak et al. 2016
source: Albayrak et al. 2016
Workflow Overview
The method starts with single-cell sorting, typically achieved using fluorescence-activated cell sorting (FACS). Each sorted cell is lysed to generate a single-cell lysate, which is subsequently divided into two aliquots for separate analyses:
- Digital Proximity Ligation Assay:
- Used to detect and quantify individual protein molecules.
- Oligonucleotide-conjugated antibodies are employed, enabling specific detection at the femtomolar level.
- RT-ddPCR:
- Used to measure mRNA expression levels.
- Ensures absolute quantification of RNA molecules at a single-cell resolution.
Both aliquots are then partitioned into droplets, allowing for absolute quantification and resulting in reliable detection of both RNA and proteins within the same cell.
This dual quantification technique is especially valuable for single-cell analysis, providing a tool to study gene expression and protein synthesis dynamics simultaneously, which is critical for applications in cell biology and even cancer research. The study marks a significant advancement in the ability to perform multiplexed, quantitative single-cell measurements.
A similar, dPLA approach by Abasıyanık et al., 2020 was employed to detect bacterial genes (16S) together with a sepsis-specific, inflammation-mediating effector proteins, such as IL-6 and TNF alpha.
In this study, the authors utilized amplitude multiplexing to distinguish between 16S rRNA genes specific to gram-positive (GP) and gram-negative (GN) bacteria while simultaneously quantifying IL-6 levels in the same reaction well, creating a triplex assay. Furthermore, the study demonstrated how ddPCR could effectively correlate bacterial burden and inflammatory cytokine expression, offering a robust platform for monitoring infection dynamics and host immune responses in septic patients.
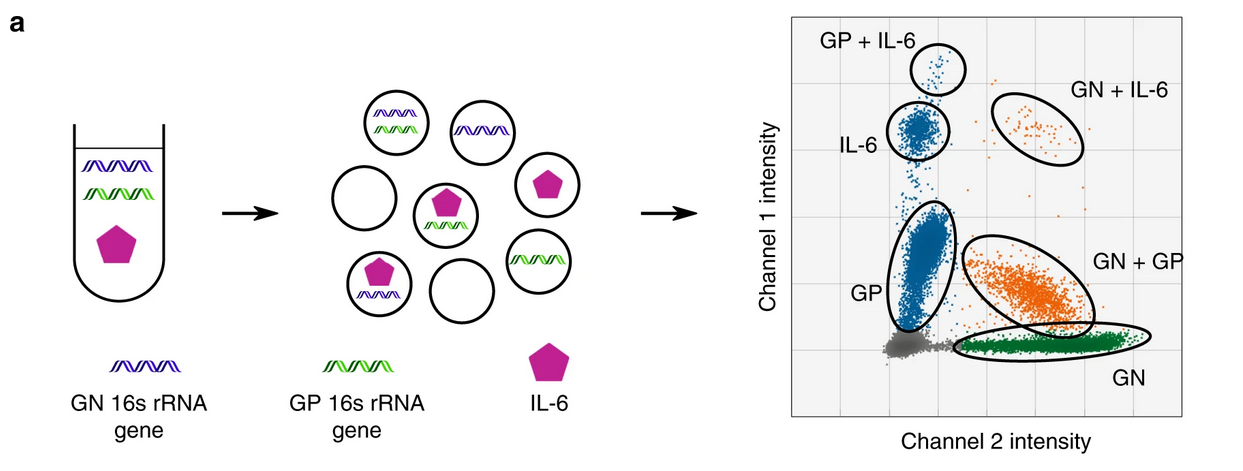 source: Abasıyanık et al., 2020 Figure 2 a
source: Abasıyanık et al., 2020 Figure 2 a
Workflow Overview
The triplex assay preparation and execution in this study was significantly more straightforward compared to the study by Albayrak et al. 2016. The workflow consisted of the following steps:
- Sample preparation and incubation:
- A few microliters of the pre-diluted (in a 0.1x TM buffer) lavage fluid sample was with proximity probes and incubated at 37°C for 1 hour.
- Addition of the reaction mix:
- This mixture was combined with a 20 µL ligation/ddPCR reaction mix containing primers, probes, and ligation additives.
- Partitioning and ddPCR:
- The final mixture was emulsified (partitioned) and subjected to thermal cycling: ligation at 25°C for 20 minutes, initial denaturation at 95°C for 5 minutes, 40 amplification cycles (94°C for 30 seconds, 60°C for 1 minute), and a final step at 98°C for 10 minutes.
This study highlights the power of ddPCR for multiplexed detection of pathogens and protein biomarkers, paving the way for improved diagnostic tools in infectious diseases and inflammatory conditions.