Singleplex/Multiplex ddPCR Reaction Optimization
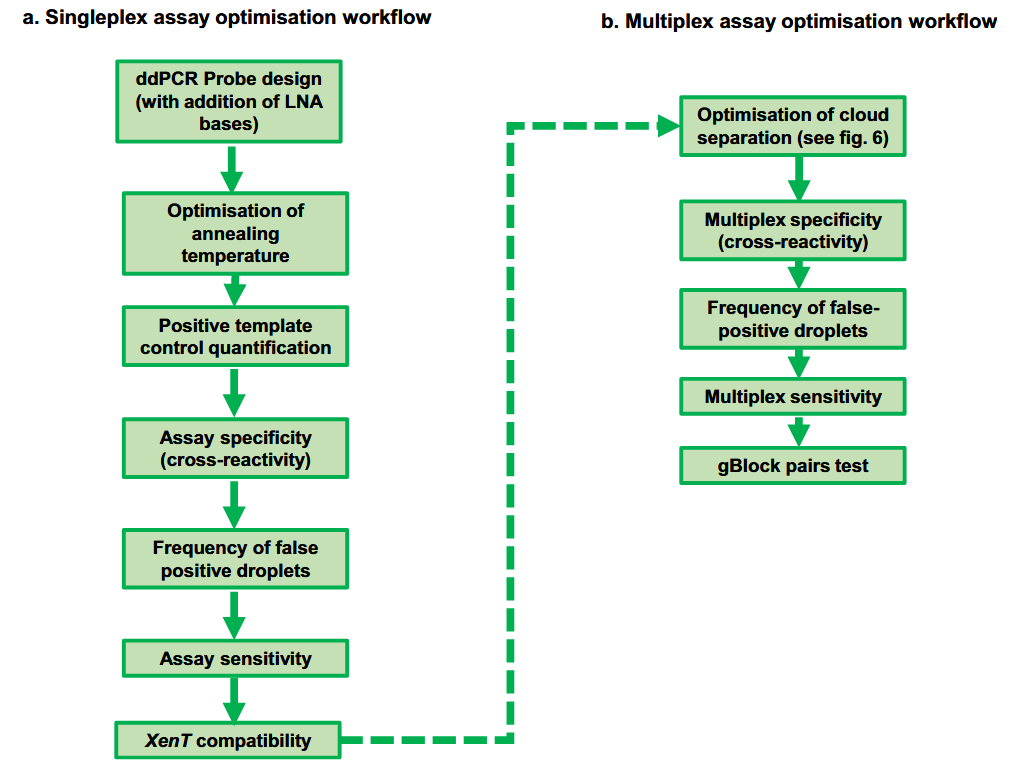
Singleplex Reaction Optimization
When you first receive your primer and probe stocks, it’s crucial to plan your initial experiments carefully. While it may be tempting to immediately test your new primer and probe assay on actual samples, optimizing the assay first is highly recommended. A great reference for ddPCR assay optimization is the study by source: Rowlands et al., 2019.
Pre-optimization Considerations
Before starting any optimization or experimental work, consider incorporating Locked Nucleic Acids (LNAs) into your probe designs. LNAs enhance probe binding strength, increasing the melting temperature (Tm). This allows for:
- Greater Tm differences between mutant and wild-type probes, reducing cross-reactivity.
- Shorter, more efficient probe designs.
For a detailed description of LNAs, see the Locked-Nucleic Acids section.
Optimization Steps
-
Annealing Temperature Gradient:
Begin by running an annealing temperature (Ta) gradient, typically between 55–65°C, to determine the temperature at which the positive and negative droplet clusters are most distinctly separated.
-
Positive Control Dilution Series:
Test a positive control, such as a gBlock or recombinant plasmid, at various concentrations. Assess cluster separation, droplet rain, and quantification robustness across replicates and dilutions.
-
Assay Specificity:
For mutation detection or species identification assays, evaluate specificity by testing the assay against similar mutations (in the same loci) or closely related species/individuals (within the same genus).
-
False Positive Rate (FPR):
Run multiple wild-type control samples to observe false positive droplets in the alternate channel. Calculate the FPR as the number of false positives per total wild-type copies. For instance, if you find 1 false positive droplet in 200,000 wild-type copies, the FPR is 0.000005%.
-
Assay Sensitivity:
Assess sensitivity by:
- Detecting a small number of target copies (e.g., 5) in increasing concentrations of background DNA (e.g., 0, 2000, 5000 copies).
- Performing a serial dilution of target DNA in a high background DNA context. Multiple replicates should be used to ensure consistent, robust results. For guidance on establishing Limits of Blank, Detection, and Quantification, see Establishing Limit of Blank/Detection/Quantification for Your Assay.
-
Carrier DNA Testing: (Optional)
If you plan to use carrier DNA (e.g., XenT), test for potential cross-reactivity between the assay and the carrier DNA.
 source: Rowlands et al., 2019, Figure 3
source: Rowlands et al., 2019, Figure 3
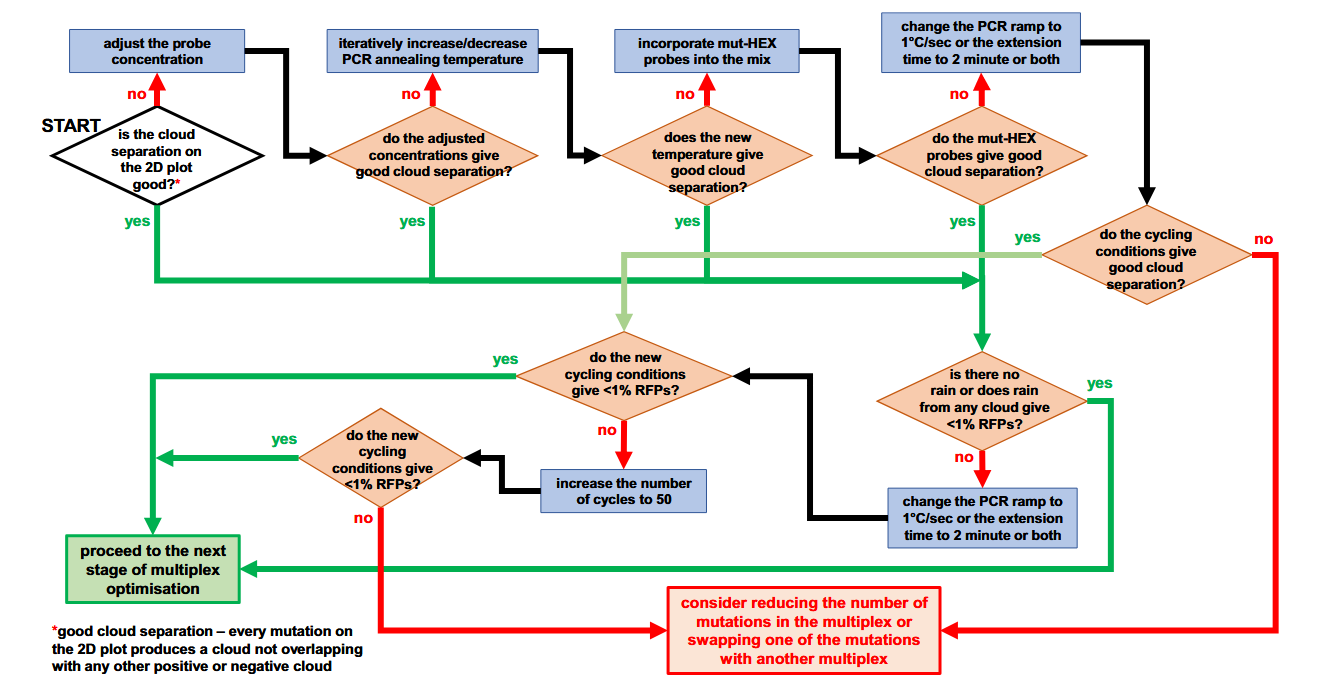
Multiplex Reaction Optimization
Before optimizing a multiplex reaction, ensure all individual assays have been optimized in singleplex. Droplet cluster separation is critical in multiplex formats, and the following workflow can help improve it:
Optimization Steps
-
Adjust Primer and Probe Concentrations:
Start with 900 nM primers and 250 nM probes, adjusting as needed.
- Iteratively Optimize Annealing Temperature (Ta).
-
Incorporate Multiple Fluorophores:
For example, include both FAM and HEX-labeled probes to distinguish targets.
- Modify PCR Conditions:
- Set the ramp rate to 1°C/second.
- Extend the annealing time to 2 minutes.
-
Increase Cycle Number:
Adjust up to 50 cycles to improve droplet cluster separation and reduce rain.
 source: Rowlands et al., 2019, Figure 6
source: Rowlands et al., 2019, Figure 6
Once the droplet clusters are optimally separated on a 2D plot:
-
Evaluate Cross-reactivity:
Test for unintended detection of unrelated mutations or targets. If no unrelated targets exist, this step can be skipped.
-
Estimate False Positive Droplets:
Run wild-type samples in multiplex format to calculate FPR for each quadrant or detection channel.
-
Assess Sensitivity:
Test the multiplex assay’s ability to detect small numbers of target copies (e.g., 5) in the presence of wild-type DNA. Focus on one mutation/target and wild-type DNA per well for simplicity.
-
Test gBlock Pairs:
Finally, evaluate how multiple targets (e.g., two mutations) behave in a single well. Check the 2D plot for artifacts before analyzing clinical or experimental samples.
By following these guidelines and referring to the original publication, you can ensure your assays are well-optimized, producing reliable and robust results every time!