Mediator Probe PCR
The MP PCR, described by Lenhert et al., 2018 is the qPCR version of the Mediator Probe ddPCR method. In their 2018 paper, the authors describe the optimization process to improve SNR (Signal-to-Noise Ratio) for contact-quenched, fluorogenic universal reporter oligonucleotides and apply the Mediator Probe-based real-time detection principle to study acute lymphoblastic leukemia marker sequence variations. The authors also developed a set of guidelines for designing contact-quenched, fluorogenic universal reporter oligonucleotides with optimized SNRs that may also be beneficial for SNR optimization in other fluorescence-based assays that utilize contact-quenched oligonucleotides or secondary reporter molecules.
The principle of MP PCR is similar to that of MPddPCR in that it’s a two-step detection process:
- The assay uses a primary mediator probe that is label-free and sequence-specific. When the target DNA sequence is present, the polymerase (typically a thermostable DNA polymerase with 5’ exonuclease activity, such as Taq polymerase) cleaves the mediator probe during amplification. This cleavage releases a short mediator sequence into the reaction.
- The released mediator sequence then hybridizes to a universal reporter oligonucleotide designed to fluoresce upon interaction.
The universal reporter is a contact-quenched probe, meaning it consists of:
- A fluorophore
- A quencher
Upon the binding of the mediator sequence and its subsequent extension by the polymerase, the reporter undergoes a conformational change that separates the fluorophore and quencher, leading to an increase in fluorescence. The intensity of this fluorescence is directly proportional to the amount of amplified target DNA, allowing quantitative detection of the target sequence in real-time.
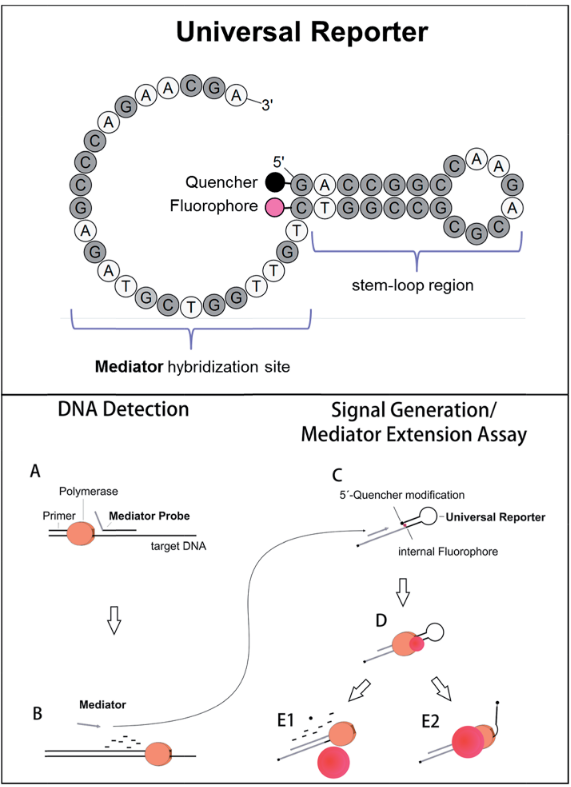
source: Lenhert et al., 2018, Figure 1