Allelic Imbalance for Copy Number Determination
Typical copy number variation (similarly to gene expression) experiments carried out with dPCR require you to run a stable reference gene assay (most often in parallel) to compare the obtained target copy numbers to a stable background. In such a way, duplications or deletions can be determined by using a simple formula a (target copy number) / b (reference copy number).
Other approaches, such as that proposed by Nell et al., 2025 can also be employed to estimate copy number alterations for particular target sequences or genes. This method relies on estimating allelic imbalance at a heterozygous germline SNP located within the target region, instead of relying exclusively on a separate genomic reference — it is therefore, essentially, a SNP-based digital PCR approach. See the comparison below between the two approaches - classical (using a reference gene locus) and SNP-based (using allelic imbalance).
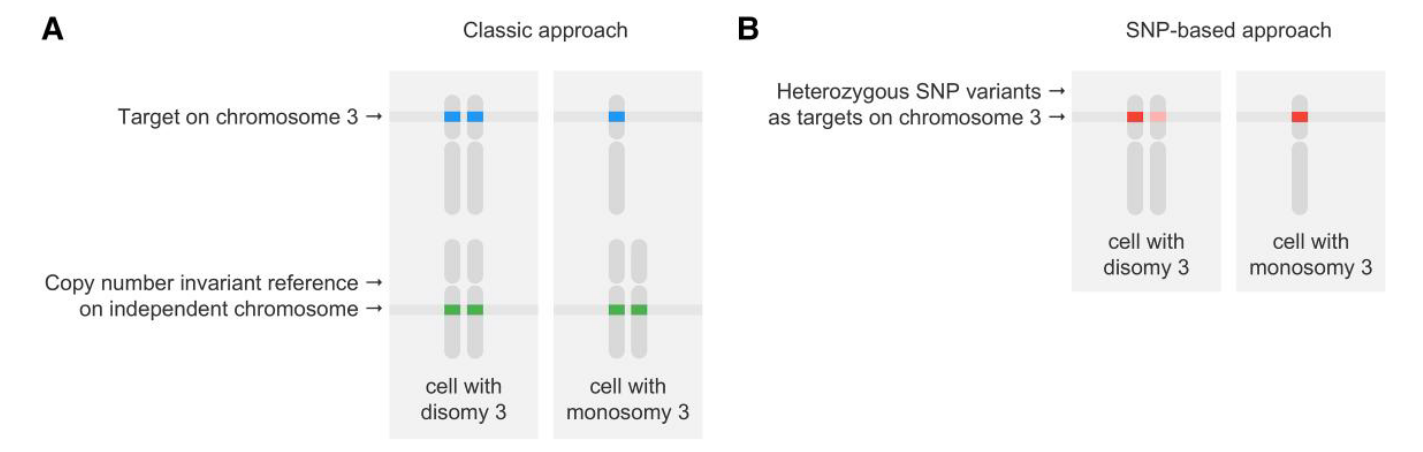
source: Nell et al., 2025, Figure 1
This SNP-based approach for determining copy number changes in the genome can be particularly useful when identifying and selecting a stable reference gene locus becomes difficult. The clear disadvantage with this approach, however, stems from the fact that heterozygous SNPs are not always available in the target locus of interest. Nevertheless, Nell and colleagues were able to show that once such assays are established, they can be significantly more precise when compared to the “classical” (target gene & reference gene) approach.